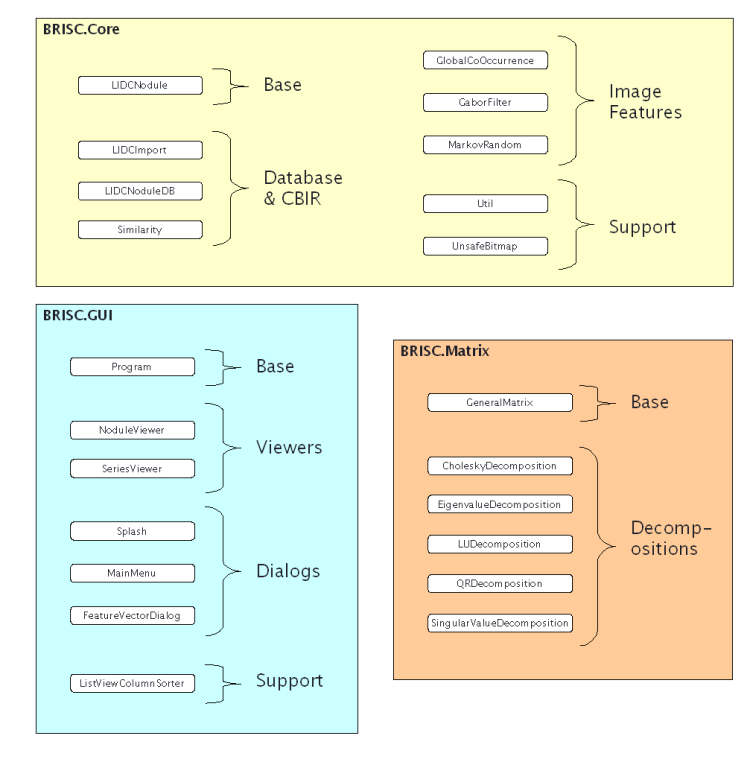

We have split our project into two namespaces: Core and GUI (there is also a Matrix namespace, but this is third-party matrix code). We have tried to keep all of our interface-specific code in GUI and all algorithm and data structure code in Core. Theoretically, it should be possible to take the classes in Core and build an entirely new CBIR interface from them, apart from the classes in GUI.
The purpose of the code in the Core namespace is to allow retrieval of Lung CT nodule images. The nodules come from the Lung Image Database Consortium (LIDC) in the form of DICOM images and XML data. The DICOM images are full CT scans of the chest. The XML contains the pixel locations of every nodule in the lung images. Each nodule has been annotated by radiologists with nodule charactaristics: calcifictation, internal structure, lobulation, malignancy, margin, sphericity, spiculation, subtlety, texture.
Due to licensing issues we will not be distributing the images or data with BRISC; however, they can be downloaded for free from http://ncia.nci.nih.gov/. Since the data is not in a format that plays nicely with BRISC, it will need to be converted to something more useful. This is taken care of by the LIDCImport class, which contains routines that read the LIDC files and produce series (.srs) files as well as all the needed nodule database XML files.
The most fundamental class in BRISC is LIDCNodule. Each LIDCNodule object represents a nodule image. Each nodule image has fields such as pixel location in the CT image and radiologist's annotations. This class also contains methods to load from and write to XML files.
All nodules information is saved and kept in XML files. The data is kept in multiple files for efficency. All of these files are create by the LIDCImport class.
The 'nodules-primary.xml' file contains the base information that is needed for each nodle. Filenames are stored in here along with slice numbers and other identifiers. Also, the pixel location in the original lung image is stored here.
The 'nodules-haralick.xml', 'nodules-gabor.xml', and 'nodules-markov.xml' files store the texture descriptors used for similarity retrieval. These can be loaded as needed so if we do not need haralick descriptors we simply do not load 'nodules-haralick.xml'. They are created by calling the appropriate methods in GaborFilter, MarkovRandom, and GlobalCooccurrence on a database of LIDCNodule objects and then saving the database to XML.
The 'nodules-points.xml' file contains the x,y coordinates for each pixel in the nodule contour. These point differ from the pixel locations in 'nodules-primary.xml' in that the primary XML file defines a rectangle around the nodule while the points XML file gives a precise boundary around the entire nodule. This data is used in the co-occurrence calculations.
The 'nodules-annotations.xml' file has the annotation data about each nodule. These are the annotations radiologists have given each nodule. Since there were four radiologists annotating each nodule there can be up to four different instances of each unique nodule.
Below is a very short, functional overview of what each class does. See the main documentation for details about the members of each class.


Handles the importing of LIDC data into LIDCNodule objects, and then the creation of .srs and .xml files for the series and nodule viewer.
This is the class that holds data about a single "nodule." Note that a LIDCNodule is unique to series, slice, nodule and physician. So, you will have multiple LIDCNodule objects for a single "nodule." Perhaps they will be from different slices, or annotated by different physicians. This class also contains structures for storing image feature data for Harlick, Gabor and Markov, as well as the PhysicalSize structure for storing actual nodule sizes in millimeters. It also contains functions for reading and writing XML.
Basically encapsulates a collection of LIDCNodule objects, providing functions to read/write the entire database from/to XML. It also contains the two core CBIR functions: RunQuery() and CalcMeanPrecisionAndRecall(). These functions run queries on the database and calculate query performance, respectively. LIDCNoduleDB also has a function to normalize Haralick features, since this uses min/max information from the entire collection (and thus cannot be done in the LIDCNodule class itself).
CBIR has two main parts: feature extraction and feature comparison. This class contains all the code for feature comparison; ie. comparing the feature vectors and/or histograms from two different nodules to see how similar they are.
These classes perform the number crunching for calculating image features. Each class contains a void-return static function that takes only one argument: an LIDCNodule object. The function calculates all image features of its type for that nodule and stores the data in the LIDCNodule structure. Note that GlobalCoOccurrence uses a small helper class CombineCoOccurrence to store temporary co-occurrence matrix data.
Contains miscellaneous stuff. Highlights include functions for loading DICOM pixel data and then converting that pixel data to .NET Bitmap objects (also handles all intensity windowing). Also contains a member DATA_PATH that is used to tell the program where the LIDC data is. This can be set from a file called "datapath.txt" in the same directory as the executable (the function LoadDataPath() reads the path from this file and is called by Program at startup).
This class encapsulates a .NET Bitmap and contains functions for locking and unlocking pixel data for unsafe access. This provides much faster pixel manipulation for the DICOM reading and windowing. This class contains a child data structure, PixelData, that is just a 24-bit color structure with 8-bit RGB fields.
Launching point for our demo app. Change the MODE constant to set program behavior. MODE=1 is an old setting and no longer supported. MODE=2 starts the GUI at the main menu. MODE=3 starts the program in task mode, in which the program performs a certain, pre-scripted set of actions, such as recalculating all Haralick features. Which task is to be performed is currently set by commenting/uncommenting lines in the doTask() function.
The two GUI viewers for our app, one for nodule data and the other for series data.
Dialogs that perform one function or another in the program.
A multi-use sorter class for ordering nodules in the viewer program. Can sort in four ways: 1) case-sensitive string, 2) case-insensitive string, 3) integer number and 4) floating- point number.
 1.4.7
1.4.7